Content Status
Type
Linked Node
Molecular Beacon Technology for GeneXpert MTB-RIF Assay
Learning Objectives-
H5Content
Content
The molecular beacons targeting the rpoB gene cover all the mutations found in more than 99.5% of Mycobacterium tuberculosis rifampicin-resistant strains.
The molecular beacon consists of:
- Loop: 18-30 base pair region of the molecular beacon, which is complementary to the target sequence
- Stem: 5-7 bp complementary sequence on both the ends of the loop
- 5’ Fluorophore: Fluorescent dye covalently linked at the 5' end
- 3’ Quencher: Nonfluorescent dye covalently linked at the 3' end
The loop structure of the molecular beacon contains the complementary oligonucleotide probe sequence, and the fluorophore and quencher molecules are attached to the ends of the stem structure (Figure 1).
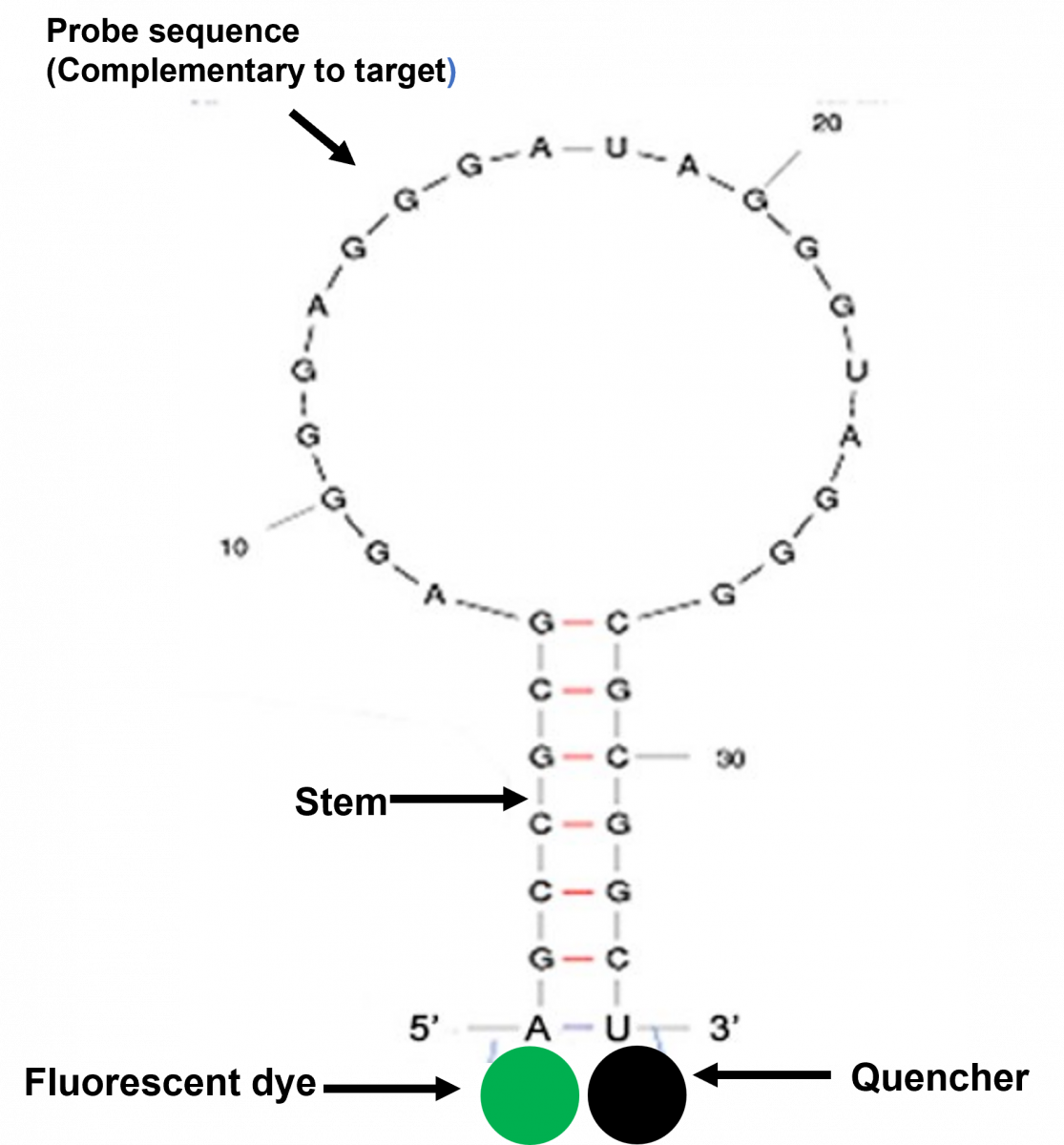
Figure 1: Structure of Molecular Beacon
Following hybridization, conformational change in the probe leads to the separation of the fluorophore and quencher molecules and the onset of fluorescence (Figure 2).
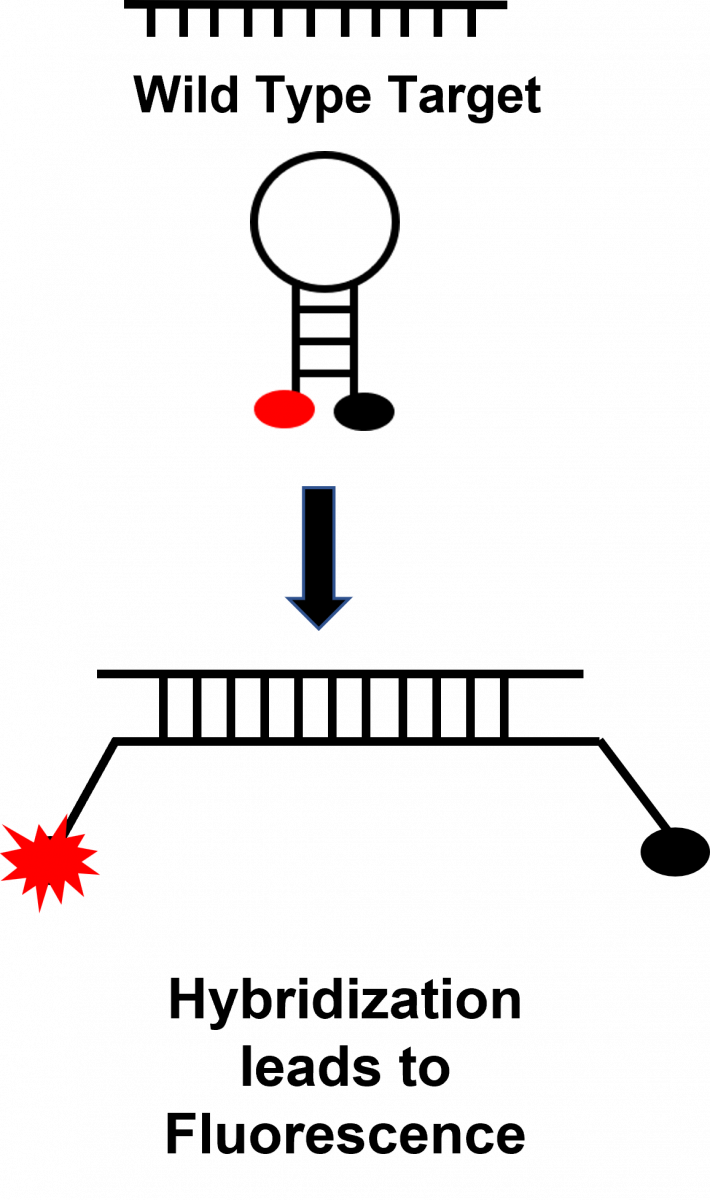
Figure 2: Fluorescence after Hybridization
Resources
Kindly provide your valuable feedback on the page to the link provided HERE
LMS Page Link
Content Creator
Reviewer
- Log in to post comments