Content Status
Type
Linked Node
Interpretation of SL-LPA: Predicting Drug Resistance
Learning Objectives-
Zone developed/ not developed (failed) in Wild Type (WT) and Mutation (MUT) probes are used to predict drugs resistance in Second Line - Line Probe Assay (SL - LPA).
For Fluoroquinolones (FLQ)
Tables 1 and 2 show mutations in gyrA, gyrB genes and corresponding WT, MUT and responsible codons and mutations:
- gyrA WT1-3: gyrA wild type probes; gyrA MUT1-2, 3A-3D: gyrA mutation probes
- gyrB WT: gyrB wild type probes; gyrB MUT1-2: gyrB mutation probes
Table 1: Mutations in gyrA gene and corresponding wild type and mutation band; Source: GenoType MTBDRsl Ver 2.0 Kit, Instructions for Use.
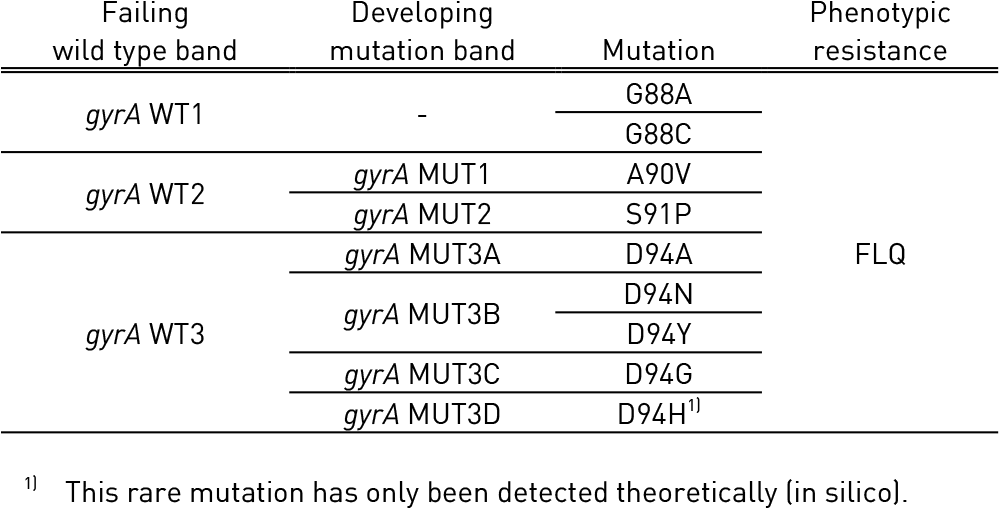
Table 2: Mutations in gyrB gene and corresponding wild type and mutation band; Source: GenoType MTBDRsl Ver 2.0 Kit, Instructions for Use.

For Second Line Injectable Drugs - Kanamycin (KAN), Amikacin (AMK), Capreomycin (CAP)
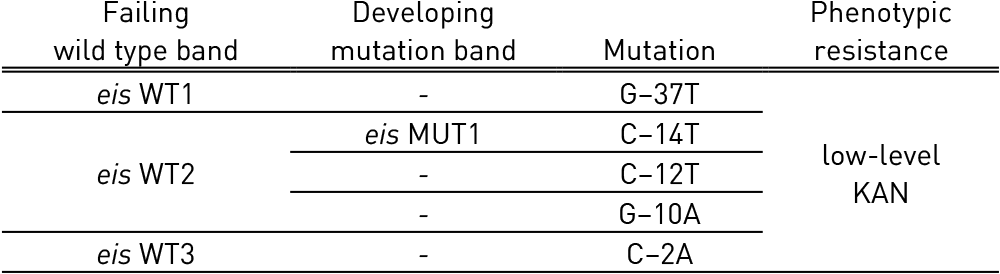
Tables 3 and Table 4 show mutations in rrs gene and eis gene respectively and corresponding WT and MUT bands:
- rrs WT 1-2: rrs wild type probe; rrs MUT 1-2: rrs mutation probe
- eis WT 1-3: eis wild type probe; eis MUT 1: eis mutation probe
Table 3: Mutations in rrs gene and corresponding WT, MUT bands and responsible codons and mutations; Source: GenoType MTBDRsl Ver 2.0 Kit, Instructions for Use.

Table 4: Mutations in eis gene and corresponding WT, MUT bands and responsible codons and mutations; Source: GenoType MTBDRsl Ver 2.0 Kit, Instructions for Use.
Resources
- GenoType MTBDRsl Ver 2.0 Kit, Instructions for Use.
- Line Probe Assays for Drug-resistant Tuberculosis Detection, GLI.
Kindly provide your valuable feedback on the page to the link provided HERE
Content Creator
Reviewer
- Log in to post comments